Unified Processing of Unstructured and Structured Data in a Lakehouse for RAG Applications
Developing Retrieval-Augmented Generation (RAG) applications within a Lakehouse architecture presents specific challenges. Integrating diverse data types like text, images, videos, and structured tables requires a robust and flexible architecture. Ensuring data quality and consistency across different formats and sources necessitates comprehensive validation and transformation processes. Managing and storing large volumes of unstructured and structured data efficiently while maintaining scalability and performance is a significant challenge. The processing and analysis of these data types together demand advanced algorithms and substantial computing power. Additionally, robust data governance, security, and compliance across various data types and sources add to the complexity.
Despite these challenges, the unified processing of unstructured and structured data is essential for RAG application development. This approach enables the integration of diverse data types, providing a holistic view and uncovering deeper insights that might not be apparent when analyzing data separately. A unified approach streamlines operations, reducing the need for multiple systems, thus simplifying maintenance and lowering operational costs. It ensures data consistency and accuracy, improving the reliability of data-driven decisions. Unified processing allows for advanced analytics, combining insights from different data types for comprehensive and actionable insights. This approach optimizes resource utilization, enhances scalability and flexibility, simplifies architecture, and improves data consistency by reducing data duplication and movement. Furthermore, a simplified architecture with unified processing reduces operational overhead, improves development efficiency, and enhances data consistency, making it crucial for effective and streamlined RAG applications.
Unified Data Pipeline Solution Overview
Data Source:
- Unstructured files on AWS S3 (PDF, Email, JPG, etc.)
AI Data Transformation:
- Transform unstructured data into JSON format, including embeddings, text summaries, and image summaries.
Data Load into Singdata Lakehouse:
- Load raw data into the
raw_table, storing various metadata and content related to files and elements.
Data Clean and Transform (Singdata Lakehouse Vector/Inverted INDEX):
- Clean and transform raw data into the
silver_table with vector index and inverted index.
Data Retrieval (Singdata Lakehouse SQL):
- Perform vector and text searches using Singdata Lakehouse SQL to retrieve and analyze data.
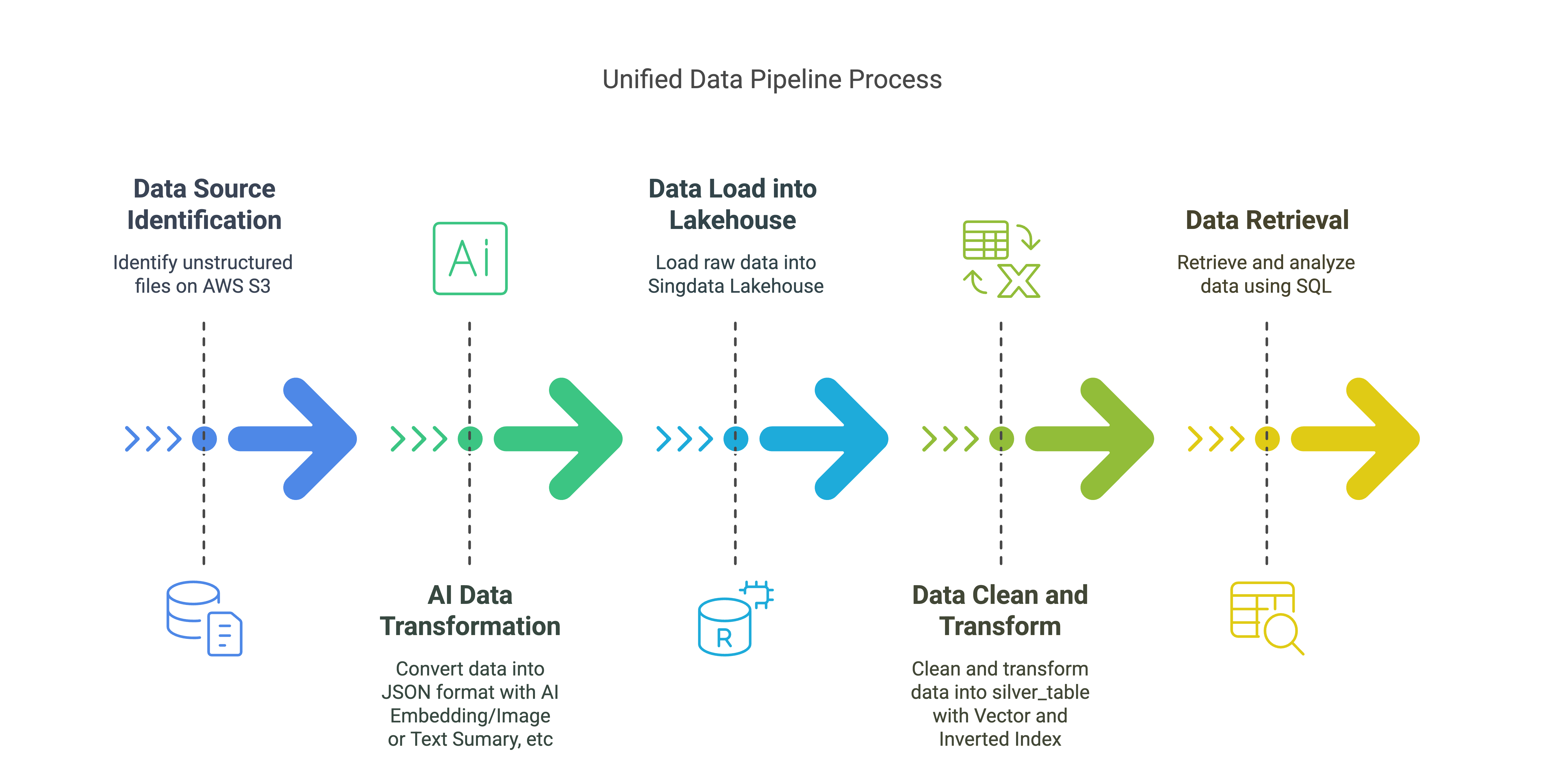
Key Components:
AWS S3: Store unstructured data
Unstructured: Ingesting Unstructured Data from S3, Transform unstructured data into JSON format
Unstructured Singdata Lakehouse Connector: Load data into Singdata Lakehouse
Singdata Lakehouse: Store and manage transformed data for RAG Application with Vector Index and Inverted Index
In this quick tutorial we'll ingest PDFs/Emails/Images from an S3 bucket in same directory, transform them into a normalized JSON with Unstructured, which we will then chunk, embed and load into Singdata Lakehouse table.
Then RAG application could retrieve the data in Singdata Lakehouse and get the embeddings and the text/graph content in a format that is ready to be used in a RAG application.
Prerequisites:
A. Get your Unstructured Serverless API key. It comes with a 14-day trial, and a cap of 1000 pages/day.
B. Get your Singdata Lakehouse Account. It comes with 1 month trial and ¥200 coupons.
C. Create an AWS S3 bucket, and populate it with PDFs of choice. Make sure to note down your credentials.
D. Install the necessary libraries:
1. Open a terminal and create new Python3.9.21 environment: unstructured
conda create -n unstructured python=3.9
conda activate unstructured
Then select unstructured as current environment
2. You could contact with qiliang@clickzetta.com to get unstructured_ingest-0.5.5-py3-none-any.whl.
You could Get the Source Code From Github Repository.
!pip install -U dist/unstructured_ingest-0.5.5-py3-none-any.whl --force-reinstall
!pip install -U "unstructured-ingest[s3, pdf, clickzetta, embed-huggingface]"
!pip install --force-reinstall "unstructured-ingest[clickzetta]"
!pip install python-dotenv
import json
import pandas as pd
import logging
import warnings
logging.basicConfig(level=logging.ERROR)
warnings.filterwarnings("ignore", category=UserWarning)
# if you want to drop the tables, set drop_tables to True
drop_tables = True
Load env variables
In this example we're loading the environment variables with all the secrets from a file in Localfile. The .evn file includes the following variables:
cz_username: Username for connecting to the Lakehouse service
cz_password: Password for connecting to the Lakehouse service
cz_service: Name of the Lakehouse service to connect to
cz_instance: Instance name of the Lakehouse service to connect to
cz_workspace: Workspace name of the Lakehouse service to connect to
cz_schema: Schema name of the Lakehouse service to connect to
cz_vcluster: Virtual cluster name of the Lakehouse service to connect to
AWS_KEY: Key for connecting to AWS services
AWS_SECRET: Secret key for connecting to AWS services
AWS_S3_NAME: Bucket name for connecting to AWS S3 service
UNSTRUCTURED_API_KEY: API key for connecting to the UNSTRUCTURED API
UNSTRUCTURED_URL: URL for connecting to the UNSTRUCTURED API
import os
import dotenv
dotenv.load_dotenv('./.env') # replace with the path to your .env file
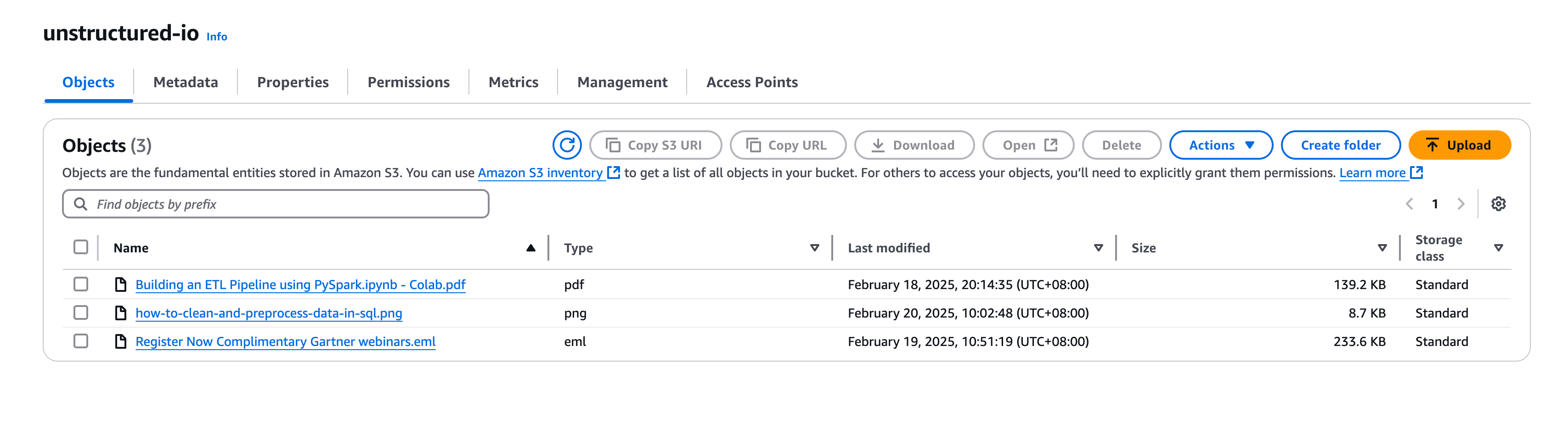
Put Unstructure Data in AWS S3
Create index in Singdata Lakehouse
Before we build the unstructured data preprocessing pipeline, let's create a Singdata Lakehouse schema and a table in it to store the processed data.
For an example of a schema, please refer to Unstructured documentation. If you'll be using the schema from the documentation, make sure that the dims value for the embeddings matches the number of dimensions of the embeddings model you choose to use. In this example it's set to 768, but your embedding model may produce vectors of a different dimension.
# Define the table names to use for storing the data in Lakehouse.
raw_table_name = "raw_elements"
silver_table_name = "elements"
embeddings_dimensions = 768
# Get the connection parameter to Singdata Lakehouse.
_username = os.getenv("cz_username")
_password = os.getenv("cz_password")
_service = os.getenv("cz_service")
_instance = os.getenv("cz_instance")
_workspace = os.getenv("cz_workspace")
_schema = os.getenv("cz_schema")
_vcluster = os.getenv("cz_vcluster")
This silver layer table is designed to store various metadata and content related to files and elements. The two indexes optimize the performance of specific queries:
The inverted text index enhances full-text search capabilities, making it easier to find records based on text content.
The embeddings vector index optimizes similarity searches on vector data, which is useful for tasks that involve comparing and finding similar elements based on their embeddings.
Benefits for RAG (Retrieval-Augmented Generation) Application Development:
Enhanced Search Efficiency: By supporting both inverted and vector searches, this table allows RAG applications to efficiently retrieve relevant information based on both text content and semantic similarity. This enhances the model's ability to find and generate contextually relevant responses.
Improved Accuracy: The combination of full-text and similarity searches ensures that RAG applications can access a broader range of relevant data, improving the accuracy and relevance of generated content.
Scalability: With optimized indexes, the table can handle large volumes of data and perform searches quickly, supporting the scalability needs of RAG applications.
Simplified Architecture: Combining inverted text and vector search capabilities in a single table eliminates the need for separate text and vector search databases. This simplifies maintenance, reduces operational overhead, and improves development efficiency.
Data Consistency: Reducing the number of data replicas from three to one enhances data consistency, minimizes data duplication, and reduces the need for data synchronization and movement.
Overall, these indexes ensure that searches and retrievals on the text and embeddings fields are performed efficiently, supporting quick and accurate query results, which are crucial for the development of effective and streamlined RAG applications.
# Define the schema to use for storing the data in Singdata Lakehouse.
raw_table_ddl = f"""
CREATE TABLE IF NOT EXISTS {_schema}.{raw_table_name} (
id STRING, -- Auto-increment sequence
record_locator STRING,
type STRING,
record_id STRING, -- Record identifier from the data source (e.g., record locator in connector metadata)
element_id STRING, -- Unique identifier for the element (SHA-256 or UUID)
filetype STRING, -- File type (e.g., PDF, DOCX, EML, etc.)
file_directory STRING, -- Directory where the file is located
filename STRING, -- File name
last_modified TIMESTAMP, -- Last modified time of the file
languages STRING, -- Document language, supports a list of multiple languages
page_number STRING, -- Page number (applicable for PDF, DOCX, etc.)
text STRING, -- Extracted text content
embeddings STRING, -- Vector data
parent_id STRING, -- Parent element ID, used to represent element hierarchy
is_continuation BOOLEAN, -- Whether it is a continuation of the previous element (used in chunking)
orig_elements STRING, -- Original element in JSON format (used to store the complete element structure)
element_type STRING, -- Element type (e.g., NarrativeText, Title, Table, etc.)
coordinates STRING, -- Element coordinates (stored in JSONB format)
link_texts STRING, -- Added field: Link text
link_urls STRING, -- Added field: Link URL
email_message_id STRING, -- Added field: Email message ID
sent_from STRING, -- Added field: Sender
sent_to STRING, -- Added field: Recipient
subject STRING, -- Added field: Subject
url STRING, -- Added field: URL
version STRING, -- Added field: Version
date_created TIMESTAMP, -- Added field: Creation date
date_modified TIMESTAMP, -- Added field: Modification date
date_processed TIMESTAMP, -- Added field: Processing date
text_as_html STRING, -- Added field: Text in HTML format
emphasized_text_contents STRING,
emphasized_text_tags STRING
);
"""
silver_table_ddl = f"""
CREATE TABLE IF NOT EXISTS {_schema}.{silver_table_name} (
id STRING, -- Auto-increment sequence
record_locator STRING,
type STRING,
record_id STRING, -- Record identifier from the data source (e.g., record locator in connector metadata)
element_id STRING, -- Unique identifier for the element (SHA-256 or UUID)
filetype STRING, -- File type (e.g., PDF, DOCX, EML, etc.)
file_directory STRING, -- Directory where the file is located
filename STRING, -- File name
last_modified TIMESTAMP, -- Last modified time of the file
languages STRING, -- Document language, supports a list of multiple languages
page_number STRING, -- Page number (applicable for PDF, DOCX, etc.)
text STRING, -- Extracted text content
embeddings vector({embeddings_dimensions}), -- Vector data
parent_id STRING, -- Parent element ID, used to represent element hierarchy
is_continuation BOOLEAN, -- Whether it is a continuation of the previous element (used in chunking)
orig_elements STRING, -- Original element in JSON format (used to store the complete element structure)
element_type STRING, -- Element type (e.g., NarrativeText, Title, Table, etc.)
coordinates STRING, -- Element coordinates (stored in JSONB format)
link_texts STRING, -- Added field: Link text
link_urls STRING, -- Added field: Link URL
email_message_id STRING, -- Added field: Email message ID
sent_from STRING, -- Added field: Sender
sent_to STRING, -- Added field: Recipient
subject STRING, -- Added field: Subject
url STRING, -- Added field: URL
version STRING, -- Added field: Version
date_created TIMESTAMP, -- Added field: Creation date
date_modified TIMESTAMP, -- Added field: Modification date
date_processed TIMESTAMP, -- Added field: Processing date
text_as_html STRING, -- Added field: Text in HTML format
emphasized_text_contents STRING,
emphasized_text_tags STRING,
INDEX inverted_text_index (text) INVERTED PROPERTIES('analyzer'='unicode'),
INDEX embeddings_vec_idx(embeddings) USING vector properties (
"scalar.type" = "f32",
"distance.function" = "l2_distance")
);
"""
clean_transformation_data_sql = f"""
INSERT INTO {_schema}.{silver_table_name}
SELECT
id,
record_locator,
type,
record_id,
element_id,
filetype,
file_directory,
filename,
last_modified,
languages,
page_number,
text,
CAST(embeddings AS VECTOR({embeddings_dimensions})) AS embeddings,
parent_id,
is_continuation,
orig_elements,
element_type,
coordinates,
link_texts,
link_urls,
email_message_id,
sent_from,
sent_to,
subject,
url,
version,
date_created,
date_modified,
date_processed,
text_as_html,
emphasized_text_contents,
emphasized_text_tags
FROM {_schema}.{raw_table_name};
"""
# Define the function to create the connection to Singdata Lakehouse.
from clickzetta.connector import connect
import pandas as pd
def get_connection(password, username, service, instance, workspace, schema, vcluster):
connection = connect(
password=password,
username=username,
service=service,
instance=instance,
workspace=workspace,
schema=schema,
vcluster=vcluster)
return connection
# Create the connection to Singdata Lakehouse.
conn = get_connection(password=_password, username=_username, service=_service, instance=_instance, workspace=_workspace, schema=_schema, vcluster=_vcluster)
# Function to execute SQL statements
def excute_sql(conn,sql_statement: str):
with conn.cursor() as cur:
stmt = sql_statement
cur.execute(stmt)
results = cur.fetchall()
return results
if drop_tables:
excute_sql(conn,f"DROP TABLE IF EXISTS {_schema}.{raw_table_name}")
excute_sql(conn,f"DROP TABLE IF EXISTS {_schema}.{silver_table_name}")
# Create Table in Singdata Lakehouse
excute_sql(conn, raw_table_ddl)
excute_sql(conn, silver_table_ddl)
Creating a database may take a few seconds. Let's check the status. We want to make sure that it says healthy before we begin writing into it.
PDFs/Images/Emails ingestion and preprocessing pipeline
Unstructured ingestion and transformation pipeline is compiled from a number of necessary configs. These don't have to be in the exact same order.
-
ProcessorConfig: defines general processing behavior
-
S3IndexerConfig, S3DownloaderConfig, S3ConnectionConfig: control data ingestion from S3, including source location, and authentication options.
-
PartitionerConfig: describes partitioning behavior. Here we only set up authentication for the Unstructured API, but you can also control partitioning parameters such as partitioning strategy through this config. We're going with the defaults.
-
ChunkerConfig: defines the chunking strategy, and chunk sizes.
-
EmbedderConfig: sets up connection to an embedding model provider to generate embeddings for data chunks.
-
ClickzettaConnectionConfig, ClickzettaUploadStagerConfig, ClickzettaUploaderConfig: control the final step of the pipeline - data loading into Singdata Lakehouse RAW table.
from unstructured_ingest.v2.interfaces import ProcessorConfig
from unstructured_ingest.v2.pipeline.pipeline import Pipeline
from unstructured_ingest.v2.processes.chunker import ChunkerConfig
from unstructured_ingest.v2.processes.connectors.fsspec.s3 import (
S3ConnectionConfig,
S3DownloaderConfig,
S3IndexerConfig,
S3AccessConfig,
)
from unstructured_ingest.v2.processes.embedder import EmbedderConfig
from unstructured_ingest.v2.processes.partitioner import PartitionerConfig
from unstructured_ingest.v2.processes.connectors.sql.clickzetta import (
ClickzettaConnectionConfig,
ClickzettaAccessConfig,
ClickzettaUploadStagerConfig,
ClickzettaUploaderConfig
)
pipeline = Pipeline.from_configs(
context=ProcessorConfig(
verbose=True,
tqdm=True,
num_processes=20,
),
indexer_config=S3IndexerConfig(remote_url=os.getenv("AWS_S3_NAME")),
downloader_config=S3DownloaderConfig(),
source_connection_config=S3ConnectionConfig(
access_config=S3AccessConfig(
key=os.getenv("AWS_KEY"),
secret=os.getenv("AWS_SECRET"))
),
partitioner_config=PartitionerConfig(
partition_by_api=True,
api_key=os.getenv("UNSTRUCTURED_API_KEY"),
partition_endpoint=os.getenv("UNSTRUCTURED_URL"),
),
chunker_config=ChunkerConfig(
chunking_strategy="by_title",
chunk_max_characters=512,
chunk_combine_text_under_n_chars=200,
),
embedder_config=EmbedderConfig(
embedding_provider="huggingface", # "langchain-huggingface" for ingest v<0.23
embedding_model_name="BAAI/bge-base-en-v1.5",
),
destination_connection_config=ClickzettaConnectionConfig(
access_config=ClickzettaAccessConfig(password=_password),
username=_username,
service=_service,
instance=_instance,
workspace=_workspace,
schema=_schema,
vcluster=_vcluster,
),
stager_config=ClickzettaUploadStagerConfig(),
uploader_config=ClickzettaUploaderConfig(table_name=raw_table_name),
)
pipeline.run()
# You could excute more SQLs to clean and transform data before insert into Silver table.、
excute_sql(conn, clean_transformation_data_sql)
Check the RAG data Ready outputs
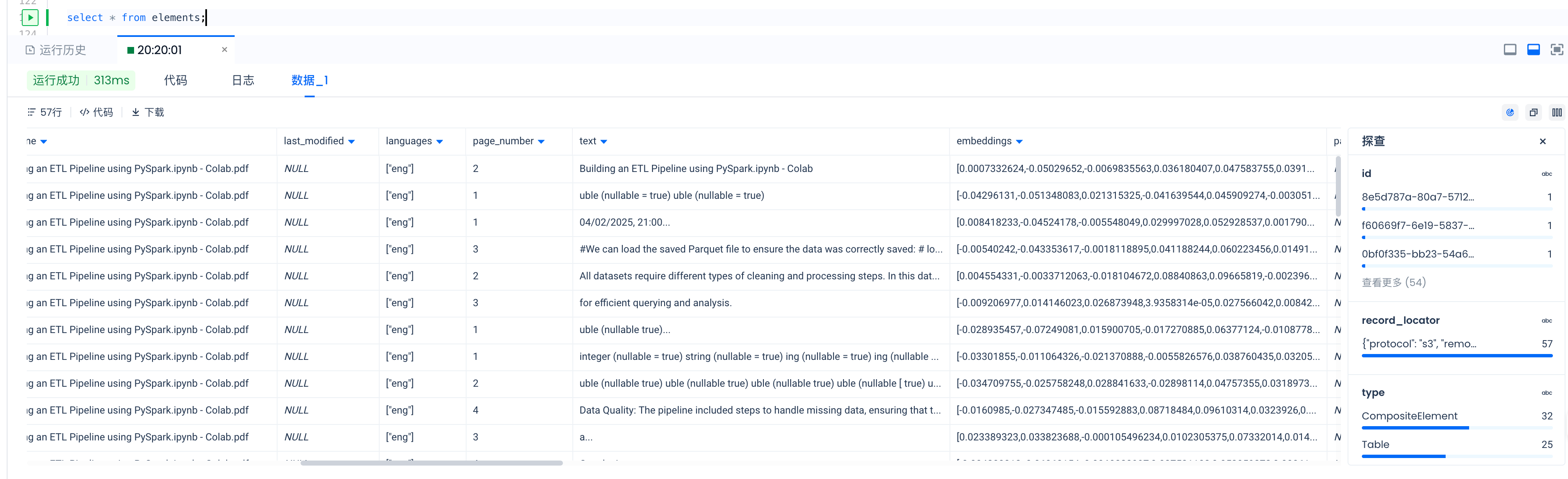
Let's connect to the Singdata Lakehouse. In the logs to the previous cell, you can see how many elements have been uploaded during the Upload Step for each document.
def get_rag_ready_data(conn, num_results: int = 5):
with conn.cursor() as cur:
stmt = f"""
SELECT
*
FROM {silver_table_name}
LIMIT {num_results}
"""
cur.execute(stmt)
results = cur.fetchall()
columns = [desc[0] for desc in cur.description] # Get column names from cursor description
rag_ready_data_df = pd.DataFrame(results, columns=columns)
return rag_ready_data_df
rag_ready_data_df = get_rag_ready_data(conn)
rag_ready_data_df

Or you could check the data Via Singdata Lakehouse Studio.
Retrieve relevant documents from Singdata Lakehouse
from sentence_transformers import SentenceTransformer
def get_embedding(query):
model = SentenceTransformer("BAAI/bge-base-en-v1.5")
return model.encode(query, normalize_embeddings=True)
def retrieve_documents(conn, query: str, num_results: int = 5):
embedding = get_embedding(query)
embedding_list = embedding.tolist()
embedding_json = json.dumps(embedding_list)
with conn.cursor() as cur:
stmt = f"""
WITH
vector_embedding_result AS (
SELECT
"vector_embedding" as retrieve_method,
record_locator,
type,
filename,
text,
orig_elements,
cosine_distance(embeddings, cast({embedding_list} as vector({embeddings_dimensions}))) AS score
FROM {silver_table_name}
ORDER BY score ASC
LIMIT {num_results}
)
SELECT * FROM vector_embedding_result
ORDER by score ASC;
"""
cur.execute(stmt)
results = cur.fetchall()
columns = [desc[0] for desc in cur.description] # Get column names from cursor description
df = pd.DataFrame(results, columns=columns)
return df
# query_text = "Harmon, Dave Scott, Bill Schmidt, Chris Teumer • Gain an action plan to hiring top IT talent • Understand how to best position yourself in the market to gain top talent • Learn why CIOs need to pay attention to hiring IT talent Register The Gartner 2025 Technology Adoption Roadmap for Infrastructure & Operations (I&O) Wednesday, February 19, 2025 EST: 10:00 a.m. | GMT: 15:00 Presented by: Ajeeta Malhotra and Amol Nadkarni • Discover why 66% of surveyed technologies are"
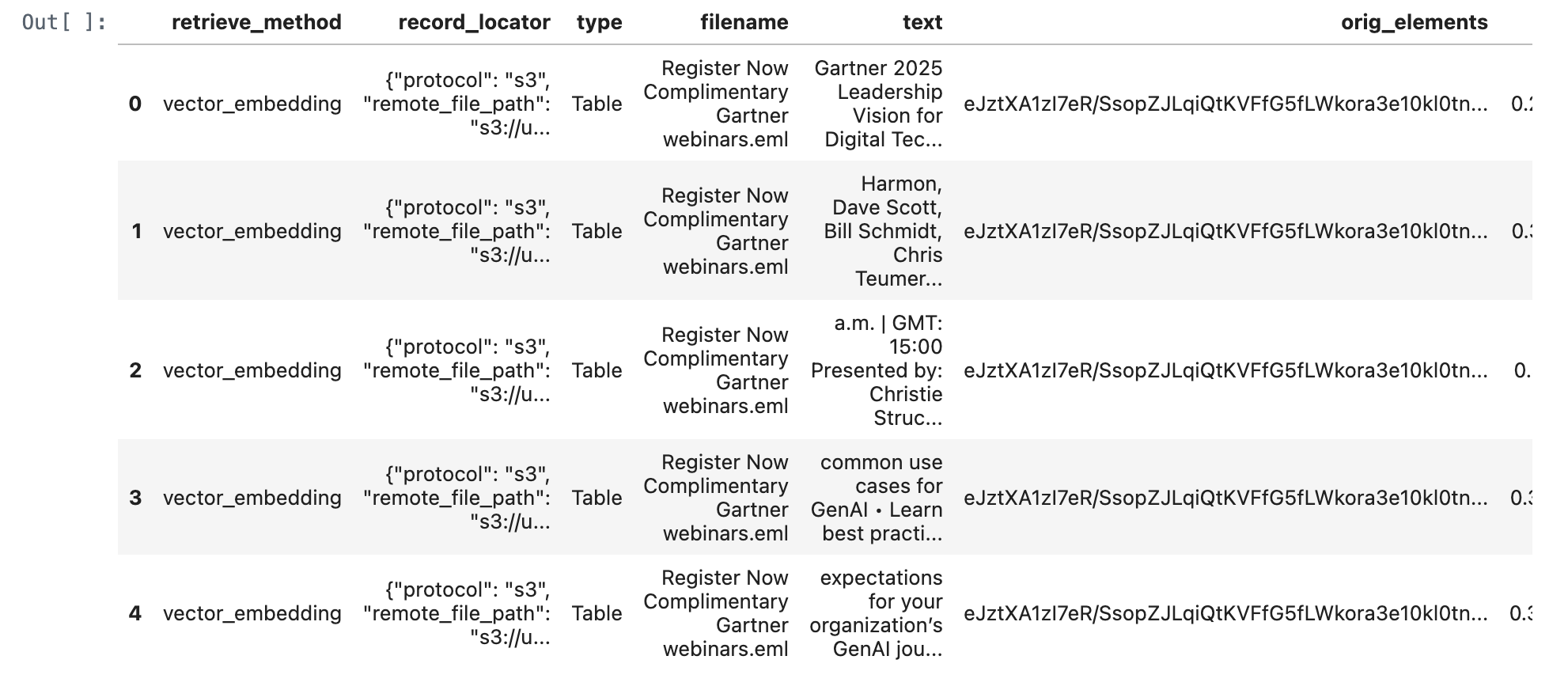
query_text = "What is gartner leadership vision for digital tech?"
retrieve_documents_df = retrieve_documents(conn, query_text)
retrieve_documents_df

def match_all_documents(conn, query: str, num_results: int = 1):
with conn.cursor() as cur:
stmt = f"""
WITH
scalar_match_all_result AS (
SELECT
"scalar_match_all" as retrieve_method,
record_locator,
type,
filename,
text,
orig_elements,
-100 AS score
FROM {silver_table_name}
WHERE match_all(
text,
"{query}",
map("analyzer", "unicode")
)
ORDER BY score ASC
LIMIT {num_results}
)
SELECT * FROM scalar_match_all_result
ORDER by score ASC;
"""
cur.execute(stmt)
results = cur.fetchall()
columns = [desc[0] for desc in cur.description] # Get column names from cursor description
df = pd.DataFrame(results, columns=columns)
return df
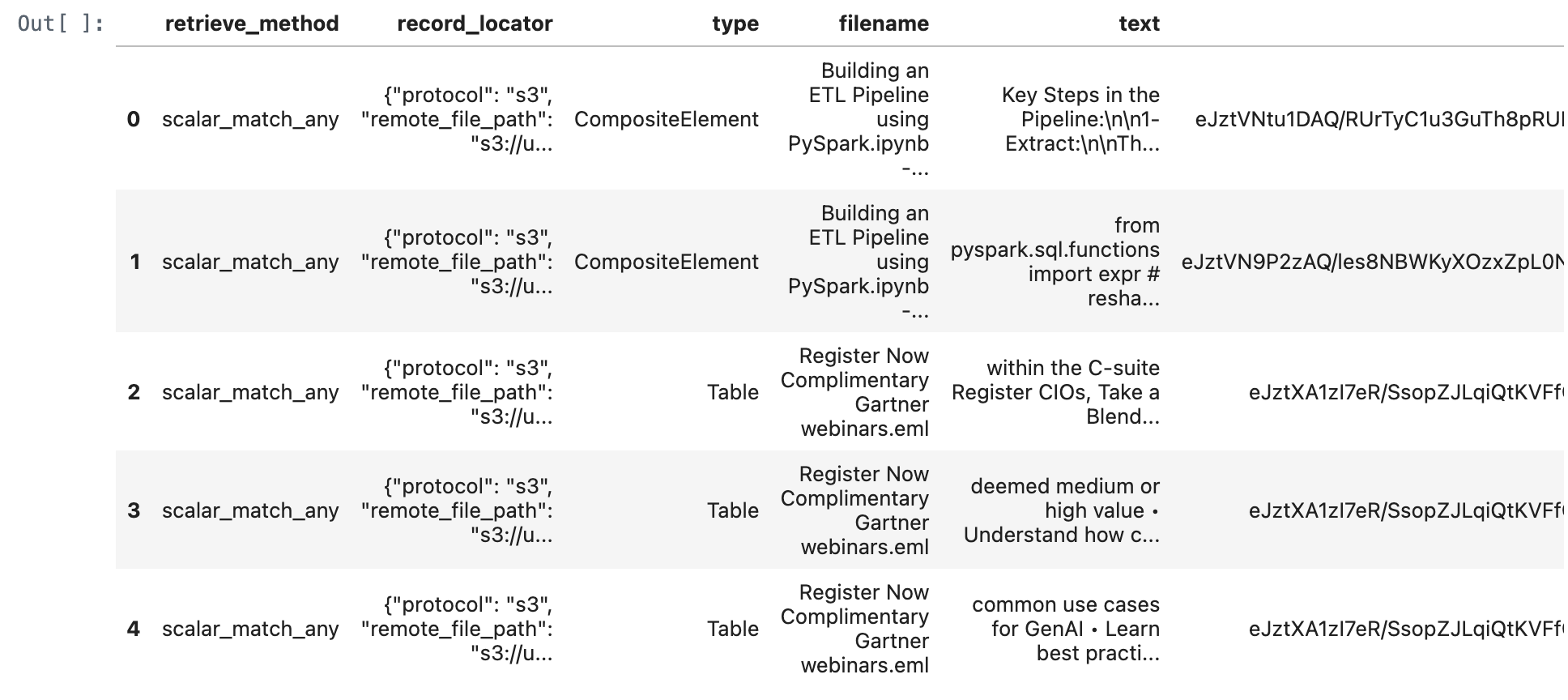
match_all_documents_df = match_all_documents(conn,query_text)
match_all_documents_df
def match_any_documents(conn, query: str, num_results: int = 5):
with conn.cursor() as cur:
stmt = f"""
WITH
scalar_match_any_result AS (
SELECT
"scalar_match_any" as retrieve_method,
record_locator,
type,
filename,
text,
orig_elements,
0 AS score
FROM {silver_table_name}
WHERE match_any(
text,
"{query}",
map("analyzer", "unicode")
)
ORDER BY score ASC
LIMIT {num_results}
)
SELECT * FROM scalar_match_any_result
ORDER by score ASC;
"""
cur.execute(stmt)
results = cur.fetchall()
columns = [desc[0] for desc in cur.description] # Get column names from cursor description
df = pd.DataFrame(results, columns=columns)
return df
match_any_documents_df = match_any_documents(conn,query_text)
match_any_documents_df
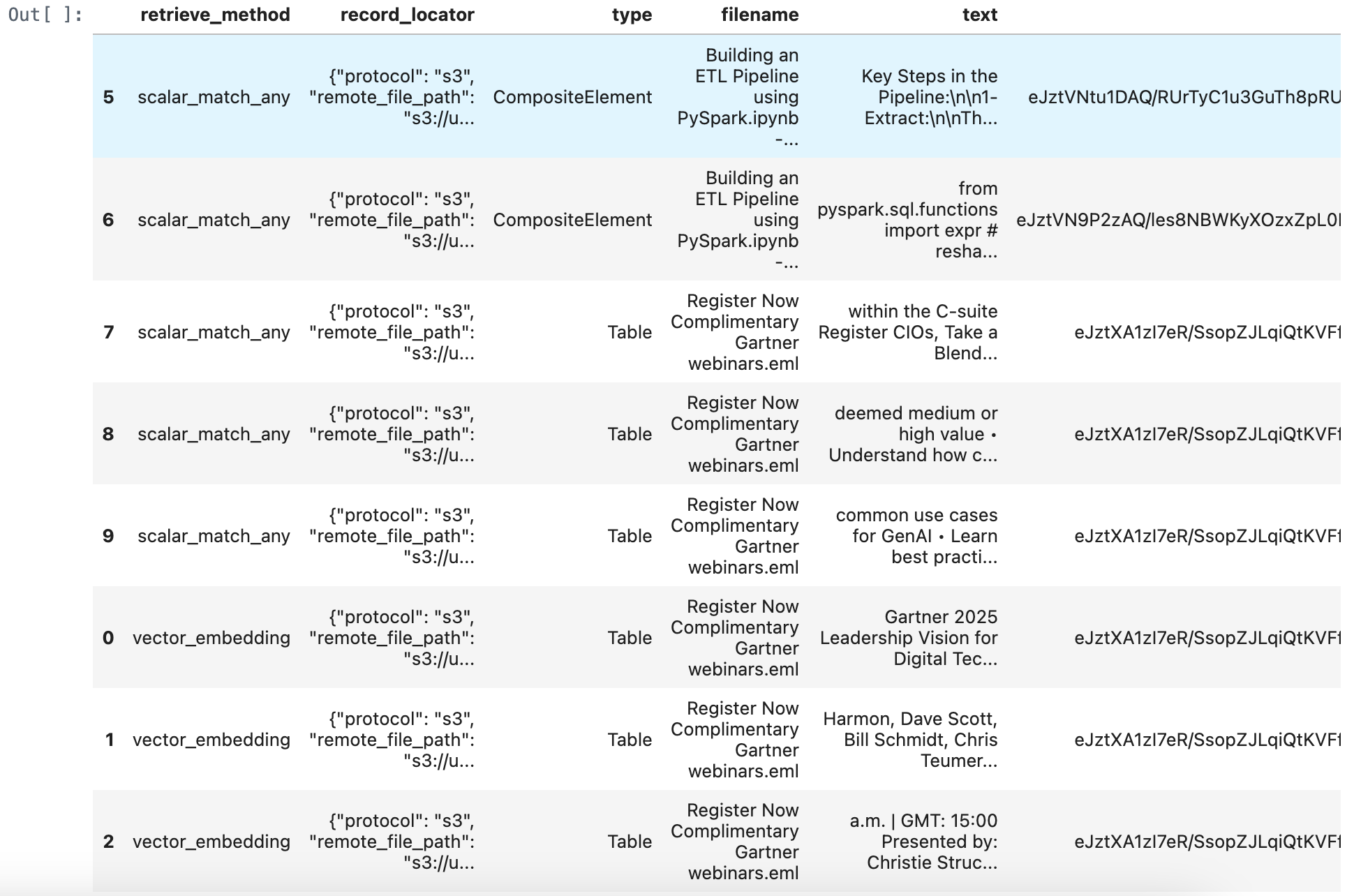
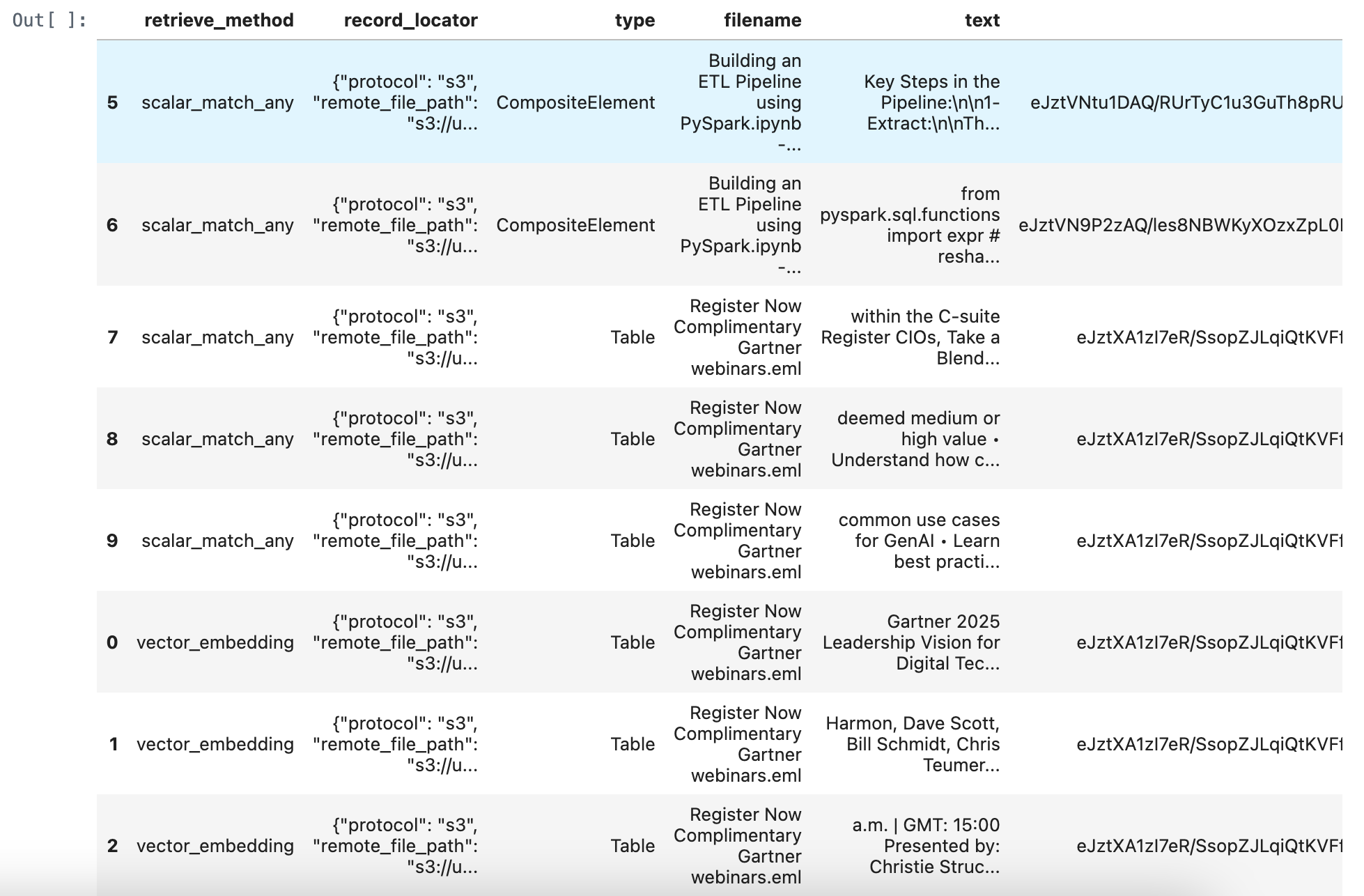
merged_df = pd.concat([retrieve_documents_df, match_all_documents_df, match_any_documents_df], ignore_index=True)
merged_df = merged_df.sort_values(by='score', ascending=True)
merged_df

import pandas as pd
import torch
import numpy as np
from transformers import AutoModelForSequenceClassification, AutoTokenizer
# Define the rerank function
def rerank_texts(query, texts, model_name="BAAI/bge-reranker-v2-m3", normalize=True):
"""
Rerank a list of texts based on their relevance to a given query using the specified reranker model.
Parameters:
- query: The query string.
- texts: List of texts to be reranked.
- model_name: The name of the reranker model to use.
- normalize: Whether to normalize the scores to the [0, 1] range using the sigmoid function.
Returns:
- A list of reranked texts.
- A list of corresponding scores.
"""
# Load the model and tokenizer
tokenizer = AutoTokenizer.from_pretrained(model_name)
model = AutoModelForSequenceClassification.from_pretrained(model_name)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device)
model.eval()
# Prepare input pairs [query, text]
pairs = [[query, text] for text in texts]
inputs = tokenizer(pairs, padding=True, truncation=True, return_tensors="pt", max_length=512)
inputs = {key: value.to(device) for key, value in inputs.items()}
# Get relevance scores
with torch.no_grad():
outputs = model(**inputs)
scores = outputs.logits.view(-1).cpu().numpy()
# Normalize scores to [0, 1] if required
if normalize:
scores = 1 / (1 + np.exp(-scores))
# Combine texts with scores and sort by score in descending order
scored_texts = list(zip(texts, scores))
scored_texts.sort(key=lambda x: x[1], reverse=True)
# Separate the sorted texts and scores
sorted_texts, sorted_scores = zip(*scored_texts)
return list(sorted_texts), list(sorted_scores)
# Example usage
# query = "Which session is presented by Ajeeta Malhotra and Amol Nadkarni?"
query = "What is gartner leadership vision for digital tech?"
sorted_texts, sorted_scores = rerank_texts(query, merged_df["text"].tolist())
# Update DataFrame with reranked texts and scores
merged_df["reranked_text"] = sorted_texts
merged_df["rerank_score"] = sorted_scores
# Get the first row of the DataFrame, which get the highest rerank_score
first_row_reranked_text = merged_df.iloc[0]['reranked_text']
print(first_row_reranked_text)
Gartner 2025 Leadership Vision for Digital Technology and Business Services Wednesday, February 19, 2025 EST: 11:00 a.m. | GMT: 16:00 Presented by: Chrissy Healey, Scott Frederick and Jennifer Barry • Revert back to growth by defining and delivering transformative impact • Resolve the asset and AI-first dilemma in delivery • Decode demand in your top accounts Register How U.S. Government Executives Can Navigate Upcoming Workforce Changes Friday, February 21, 2025 EDT: 10:00
Summary Benefits for RAG Application Development
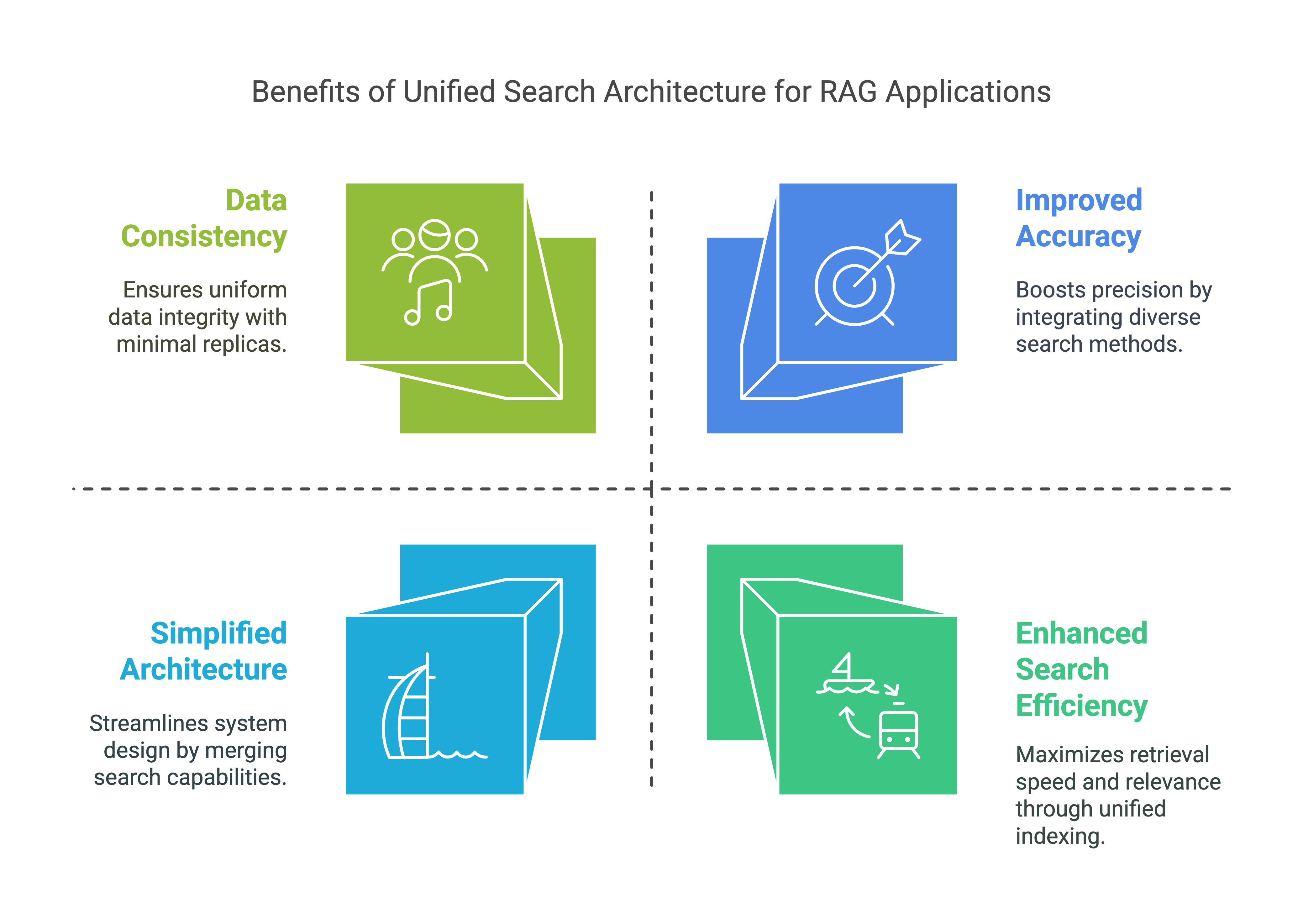
Enhanced Search Efficiency:
- By supporting both inverted and vector searches, this table allows RAG applications to efficiently retrieve relevant information based on both text content and semantic similarity. This enhances the model's ability to find and generate contextually relevant responses.
Improved Accuracy:
- The combination of full-text and similarity searches ensures that RAG applications can access a broader range of relevant data, improving the accuracy and relevance of generated content.
Scalability:
- With optimized indexes, the table can handle large volumes of data and perform searches quickly, supporting the scalability needs of RAG applications.
Simplified Architecture:
- Combining inverted text and vector search capabilities in a single table eliminates the need for separate text and vector search databases. This simplifies maintenance, reduces operational overhead, and improves development efficiency.
Data Consistency:
- Reducing the number of data replicas from three to one enhances data consistency, minimizes data duplication, and reduces the need for data synchronization and movement.
Overall, this Singdata Lakehouse architecture reduces operational complexity, enhances data consistency, and improves development efficiency, making it ideal for effective RAG application development.
Appendix:
You can also refer to github for this tour。










